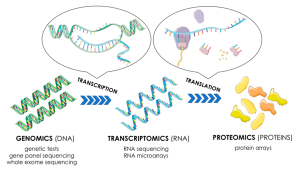
This approach is used to identify the qualitative and quantitative RNA levels in the whole genome. This includes which transcripts are present and the levels of their expression. Although only 2% of the DNA is translated in to protein, almost 80% of the genome is transcribed. This includes the coding RNA, short RNA, including microRNA, piwi RNA, small nuclear RNA. Apart from acting as an intermediate between DNA and protein, RNA also has structural and regulatory functions during native and altered states. They have been shown to have a role in myocardial infarction, adipose differentiation, diabetes, endocrine regulation, neuron development, and others. Thus, it is crucial to understand which transcripts are expressed at a time. Apart from next generation sequencing, probe-based assays, and RNA-seq are also used in this approach.
What is Transcriptomics used for?
Transcriptomics allows identification of genes and pathways that respond to and counteract biotic and abiotic environmental stresses. The non-targeted nature of transcriptomics allows the identification of novel transcriptional networks in complex systems. Transcriptome databases have grown and increased in utility as more transcriptomes are collected and shared by researchers. Measuring the expression of an organism’s genes in different tissues or conditions, or at different times, gives information on how genes are regulated and reveals details of an organism’s biology. It can also be used to infer the functions of previously unannotated genes. Transcriptome analysis has enabled the study of how gene expression changes in different organisms and has been instrumental in the understanding of human disease. An analysis of gene expression in its entirety allows detection of broad coordinated trends which cannot be discerned by more targeted assays.
Techniques
There are two key contemporary techniques in the field: microarrays, which quantify a set of predetermined sequences, and RNA-Seq, which uses high-throughput sequencing to record all transcripts. As the technology improved, the volume of data produced by each transcriptome experiment increased. As a result, data analysis methods have steadily been adapted to more accurately and efficiently analyze increasingly large volumes of data.